PhD proposal
PhD Director: Sylvain Cussat-Blanc – Institute for Research in Computer Science of Toulouse, University Toulouse
Capitole, IRIT – CNRS UMR5505, ANITI
PhD co-supervisor: Pierre Cordelier – Toulouse Cancer Research Centre, CRCT – INSERM U1037, CNRS
U5071, UPS
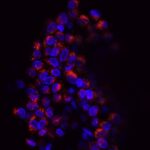
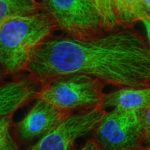
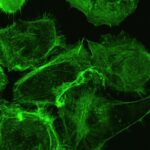
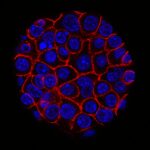
Our group within the Institut de Recherche en Informatique de Toulouse (IRIT) has developed a simulation platform that provides all the tools necessary to simulate multicellular cell microtissues growth under different environmental or treatment conditions (1). Such platform is key to easily develop models of complex biological systems by proposing various interaction paradigms such as visual programming, real-time simulation generation, simulation analysis in 2D/3D and virtual reality among others. For this PhD program, we want to model oncolytic virus infection and killing of cancer cells, to identify an optimal dose of treatment with a given schedule in silico for best therapeutic efficacy. Briefly, oncolytic virus (OV) are novel therapeutics that specifically infect, kill, replicate and propagate in cancer cells and tumors, to induce cancer cell death via multiple pathways and to recruit an immune response against tumors. Our partner team, led by Dr P. Cordelier within the cancer research center of Toulouse (CRCT), recently demonstrated that OV derived from myxoma virus (MYXV) are effective in killing primary cells derived from pancreatic cancer (PDAC), a disease with no cure (2), and are promising therapies for patients with this disease (3). However, the method and timing of administration of a given total dose of virus in a treatment cycle in complex 3D tumoroïds is still to be identified, to foster clinical valorization of this research program.
Most models of the literature are either equation-based models (4, 5, 6) or 2-D agent-based computational models (7) of the virus spreading. On the first hand, mathematical models are very precise, but they are usually complex to modify to explore various parameters on the cells. On the second hand, constraining the model to 2 dimensions takes away many parameters that can be crucial when studying the dynamics of the virus spreading. We recently generated a preliminary version of this computational model following infection with MYXV of primary PDAC cells cultured in monolayers (manuscript in preparation). With this model, we simulated cancer cell relapse following infection at a given cell confluence. Using GAGA optimization algorithm, we generated a desirable dose-schedule combination that was validated in vitro.
Because tumors are complex and heterogenous by essence, we want to move one step forward during this PhD program to design a 3D agent-based model that considers the specificities of a 3D topology (oxygen diffusion and influence on the cell cycle, etc.) while keeping the flexibility of an agent-based model, changing a cell determinant “at-a-glance”. During this project, our goal will be to model the viral replication cycle based on already available biological variables that belong to the virus and the tumour cell itself. To this end, our partner at CRCT has already collected important data for our modelling strategy. With this in hands, our objectives are (i) to analyze the impact of virus replication on the cell cycle kinetics in 3D tumoroïds, (ii) to reproduce in silico the infectious process of the viruses in their oncolytic activity, (iii) to elaborate theoretical therapeutic scenario that will be evaluated in forthcoming confirmatory biological studies. To step up to the third dimension, we will use a mass-spring-damper system [7] to simulate cellular physics, i.e., cell adhesion and collisions. This model is ready to use in the simulation platform and have been successfully validated on multicellular tumoroïds (1). Virus diffusion within the 3D tumoroids will be simulated by direct contact between cancer cells. This diffusion method is an initial hypothesis that will be challenged against biological data. Other approaches (e.g., long-range diffusion) might be considered depending on the fitting quality to these data. With such a model, we will be able to question how cell density and topology influence the virus spread and how the cell cycle is important to the killing dynamics of the virus in 3D. Addressing how cancer cells and virus coevolve in this restricted ecosystem will help test different therapeutic scenarios involving oncolytic infection in silico, with the objective to determine the best dose and frequency of administration of the virus (hitting hard or killing softly). To this end, we will use stochastic optimization (e.g., genetic algorithm) to determine the precise amount of virus to add to the cell structure at a given frequency (daily, hourly, etc.). These state-of-the-art algorithms will explore large search space with high evaluation costs of each solution. The different options will be functionally validated in vitro, then in vivo by our partner within CRCT, to determine the predictive value of the in silico model. We believe that integrating in silico modeling in preclinical research programs will help propose new strategies to improve the activity of cutting-edge oncolytic viruses for the treatment of cancer, especially for PDAC for which efficient treatments are urgently needed.
Key words: cancer, therapeutic innovation, computer modelling