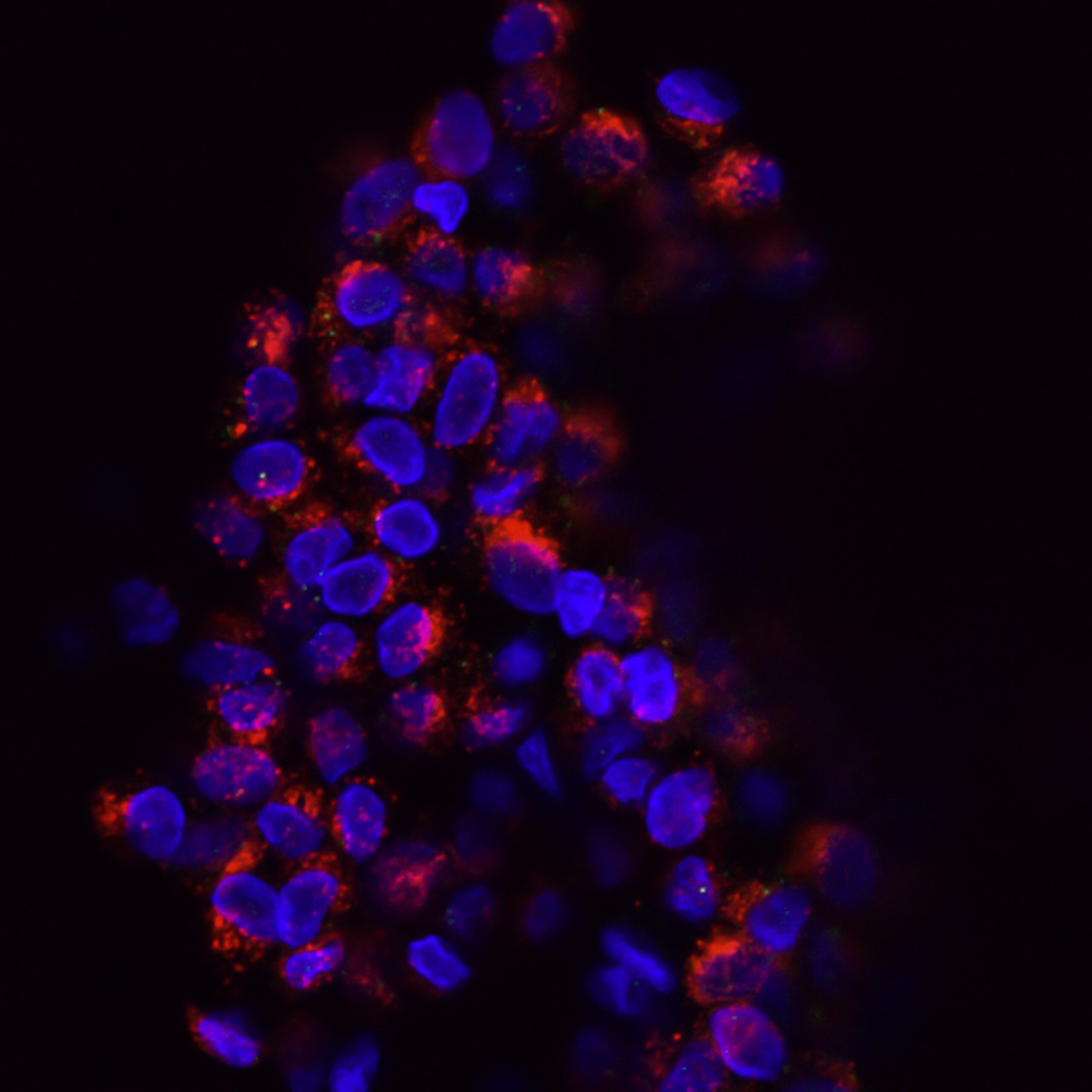
“THE ROLE OF PROPROTEIN CONVERTASES IN REGULATING TUMOR PROGRESSION, METASTASIS AND ANTITUMORAL IMMUNE RESPONSE”
Workshop: Spatial Multi-Omics for Mapping Human Tissue Function, Aging, and Diseases
On Monday, June 17, Restore lab receives Rong Fan, Ph.D. from Department of Biomedical Engineering, Yale University at 2pm for a talk about « Spatial Multi-Omics for Mapping Human Tissue Function, Aging, and Diseases« .
« Despite latest breakthroughs in single cell sequencing that revealed cellular heterogeneity, differentiation, and interactions at an unprecedented level, the study of multicellular systems needs to be conducted in the native tissue context defined by spatially resolved molecular profiles to better understand the role of spatial heterogeneity in biological, physiological and pathological processes. In this talk, I will begin with discussing the emergence of a whole new field – “spatial omics”, and then focus mainly on a new technology platform called Deterministic Barcoding in Tissue (DBiT) for spatial omics sequencing developed in our laboratory over the past years. We conceived the concept of “spatial multi-omics” and demonstrated it for the first time by co-mapping whole transcriptome and proteome (~300 proteins) pixel-by-pixel directly on a fixed tissue slide in a way compatible with clinical tissue specimens including FFPE. It has been applied to the study of developing mouse brain, human brain, and human lymphoid tissues associated with normal physiology, disease, or aging. Recently, our research enabled another new field – “spatial epigenomics” – by developing multiple DBiT-based spatial sequencing technologies for mapping chromatin accessibility (spatial-ATAC-seq), histone modification (spatial-CUT&Tag), or further combined with transcriptome or proteins for spatial co-profiling. These new technologies allow us to visualize gene expression regulation mechanisms pixel by pixel directly in mammalian tissues with a near single cell resolution. The rise of NGS-based spatial omics is poised to fuel the next wave of biomedical research revolution. Emerging opportunities and future perspectives will be discussed with regard to human biology research, aging, disease, clinical biomarker discovery and therapeutic development«
,
Biosketch
Dr. Rong Fan is the Harold Hodgkinson Professor of Biomedical Engineering at Yale University and Professor of Pathology at Yale School of Medicine. He received a Ph.D. in Chemistry from the University of California at Berkeley and completed the postdoctoral training at California Institute of Technology before joining the faculty at Yale University in 2010. His current interest is focused on developing microtechnologies for single-cell and spatial omics profiling to interrogate functional cellular heterogeneity and inter-cellular signaling network in human health and disease (e.g., cancer and autoimmunity). He co-founded IsoPlexis, Singleron Biotechnologies, and AtlasXomics. He served on the Scientific Advisory Board of Bio-Techne. He is the recipient of a number of awards including the National Cancer Institute’s Howard Temin Career Transition Award, the NSF CAREER Award, and the Packard Fellowship for Science and Engineering. He has been elected to the American Institute for Medical and Biological Engineering (AIMBE), the Connecticut Academy of Science and Engineering (CASE), and the National Academy of Inventors (NAI).

join on Teams here.
Interview of the CARe Graduate School, Philippe Valet by SANAO (association representing healthcare companies in Occitania)
Translation of the video:
What is the goal of the CARe doctoral school ?
Nowadays, Biology or research in Biology or bio-medicine research can no longer be achieved by biologists only. It’s over. That was the case when i first start to do research.
Now, we need informaticians obviously. We also need mathematicians, , physicians, chemists… At the University where I used to be Director of Masters or Director of the PhD School for a long time, tracks are quite fixed. That is to say : when you study immunology, you do immunology, when you study cancerology, you do cancerology, I used to study Pharmacology, I do pharmacology and nothing else.
I think this is over now, it cannot work that way anymore. Tracks must be mixed, merged and have to overlap. The idea of CARe is simple : gathering everybody no matter if they are researchers, as we are here in the lab « the laboratory rats », or clinician, or engineers, -lots of engineers are applying to the Care program by the way- or private companies and make everybody work together.
The CARe PhD School is a graduate school, could you explain us what it is ?
The Graduate School starts from the Master to the PhD. It is a five-year educational program that includes first year of Master, second year and PhD program.
Is it possible to go abroad ?
So during the first semester of Master 1, students stay here (in Toulouse) but they are free to go abroad as soon as the second semester. One of the main principle of CARe is to spend some internship time abroad. For a minimum of 3month to a maximum of 6 months funded by CARe-GS. If they want to stay longer, their host laboratory have to take care of the extra-funding.
How are the lessons schedulded ?
During the second year of Master, teachings are planned on the afternoons, and in English obviously. They are done in visioconference. We have a fully-connected room. A very interactive room, not just a visio link on a computer.
During the first year of Master, they have lessons on Monday, Tuesday until Wednesday noon –even if their schedule is not always full- but that is a maximum. The rest of the week they are in the labs, all year long. They are not disconnected from the labs and that is great.
What is the difference with a classic training program ?
The difference with a classic training program is that for example, Master 1 students are doing 2-months internships, Master 2 students are doing 4-monhs internships or something like that. At the CARE GS, students are doing their internship since day 1, all year long.
At the end of their training program, which diplomas do the PhD students get ?
They get a double-diploma : one from the University toulouse III- Paul Sabatier, one from the other university, from Gottingen etc. So they graduate with a PhD double diploma more a diploma supplement that is given by the CARe Graduate School, soi t is valuable on their resumes.
How many students do you have by classes ?
They are 15. Half of them are international students, CARe is funding their travel and accomodation.
Do you have any advices for high schools students ?
No matter what you are keen on, just do things you like. If it is biology, study biology, if it is mathematics, do mathematics, do something you love.
INTEGRATING MULTI-OMICS AND BIOLOGICAL KNOWLEDGE TO BRIDGE SIGNALLING AND METABOLISM
On Friday, March 22nd, Aurélien Dugourd from the Saez-Rogriguez lab, Heidelberg University will give a talk on how the integration of multi-omics and biological knowledge can bridge signalling and metabolism at 11am in Salle Cazaux, CRCT.
BIO: After he graduated from his Computational Biology master’s degree in 2015, Aurelien Dugourd joined Julio Saez-Rodriguez team as a PhD student. He worked on the development of hybrid mechanistic models, integrating gene regulation, signaling pathways and metabolomics data to explain disease phenotypes, help find new therapeutic targets and predict their potential effect based on a specific patient profile. This led to the development of the tools COSMOS and oCEan. This project was part of the collaborative SyMBioSys ITN project, financed by the European Marie Skłodowska-Curie actions, as well as the SMART-CARE consortium to apply mass-spectrometry-based systems medicine to cancer.
Today, he leads the development and application of methods to extract interpretable mechanistic insights from multi-omic datasets. He especially focuses on leveraging prior knowledge, in the context of signaling and metabolism of complex diseases, such as cancer and development of treatment resistance. He works in collaboration with pharmaceutical partners, notably Pfizer, to support the development of novel cancer treatments and better understand the development of resistances by bringing those methods closer to relevant industrial applications.

Follow the event online here.
Our 2024 call for PhD proposals is open
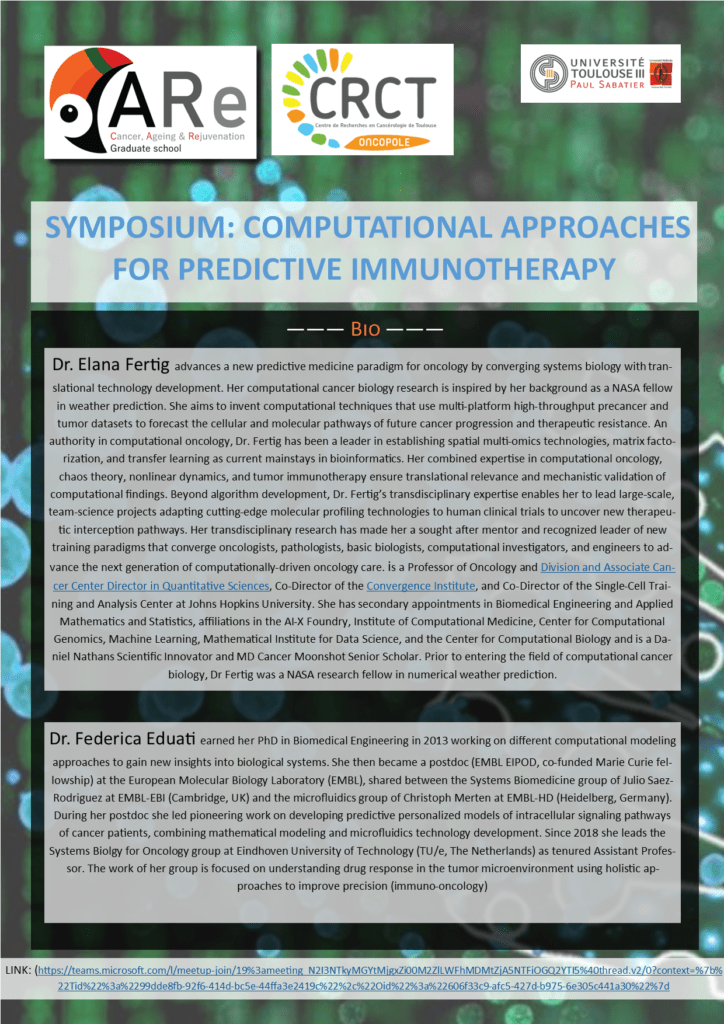
SYMPOSIUM: Computational approaches for predictive immunotherapy
Research and Technology National Agency (ANRT) « Pour un plan national en faveur du doctorat » with CARe’s Director, Philippe Valet
The conference « Pour un plan national en faveur du doctorat » (For a national plan to promote doctorates) was held on Friday 17 November at CampusCyber – La Défense, ¨Paris.
On this occasion, Sylvie Retailleau, Minister for Higher Education and Research, announced the launch of a mission on the doctorate, entrusted to Sylvie Pommier and Xavier Lazarus, which will be supported by the ANRT.
Philippe Valet, Director of the « Cancer, Ageing and Rejuvenation » Graduate School was invited to present the work and organisation of this new transdisciplinary international education program.
See Philippe Valet intervention here. (2:36:41 – 3:46:30)

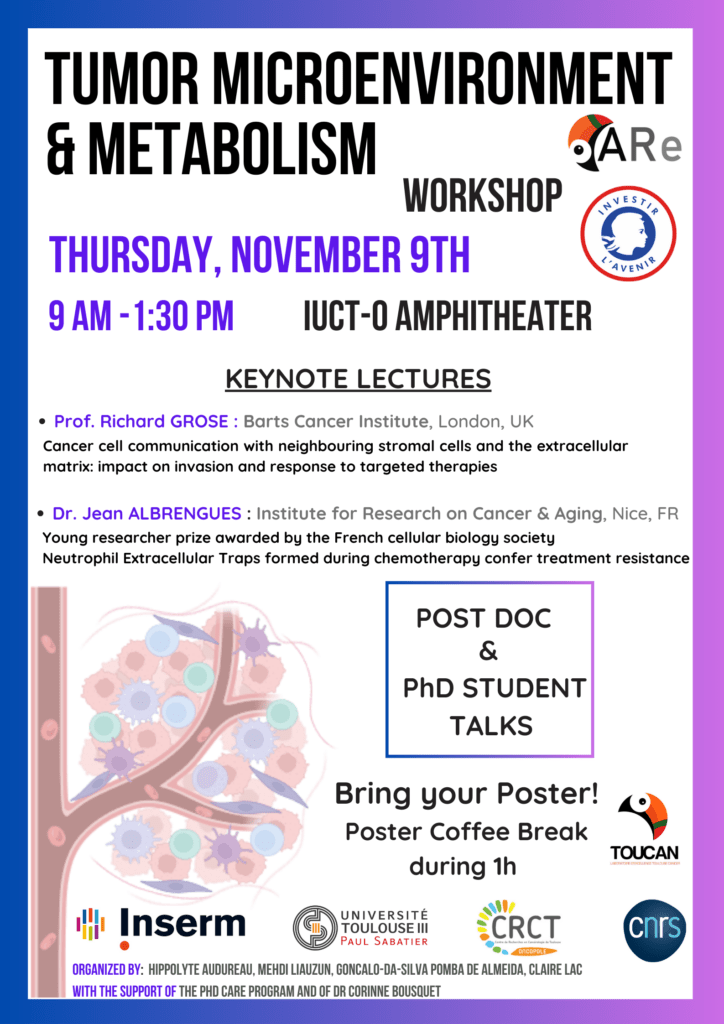
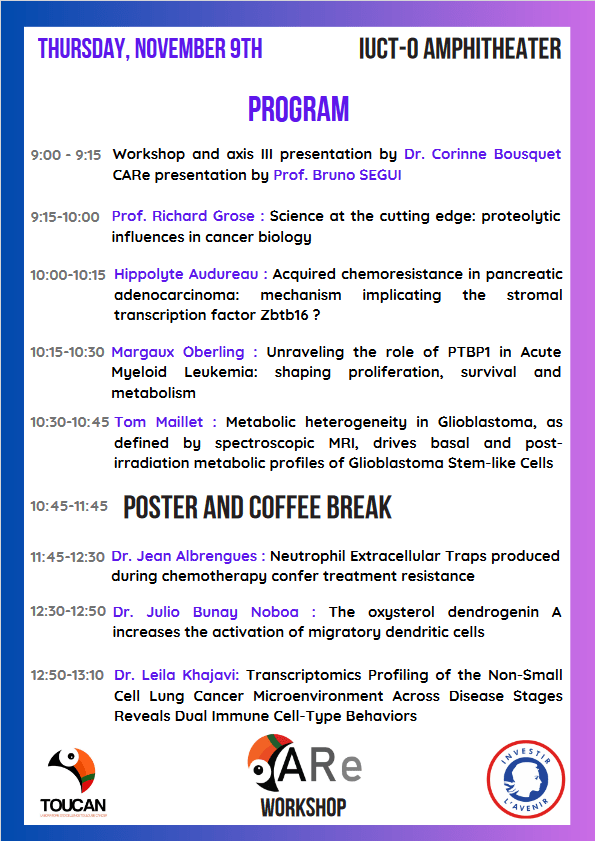
Workshop axe 3 « Tumour microenvironment and metabolism »
Insights into the disruption of gene regulatory programs in cancers – Workshop by Anthony Mathelier
On October 20 from 11 to 12, Dr Anthony Mathelier from the Molecular Medecine Norway Center will give a workshop at the Toulouse Research Center in Cancerology
Abstract:
« Most cancer somatic alterations occur in the noncoding portion of the human genome, which contains important cis-regulatory regions acting as genetic switches to ensure gene expression occurs at correct times and intensities in correct tissues. However, the identification of critical noncoding cancer driver events has been mostly limited to a few examples with high recurrence or high functional impact. Transcription factors (TFs) are key proteins binding to cis-regulatory regions at their TF binding sites to modulate the rate of gene transcription. As cancer is a disease of disrupted cellular regulation, it is critical to analyze these regions to highlight patient somatic mutations and epigenetic modifications altering the gene regulatory program of the cells. In this talk, I will present our recent works on the interplay between TF binding, somatic mutations, and DNA methylation alteration that shifts the gene regulatory program in patients cancer cells.«

If you are willing to attend this event online, please join here.